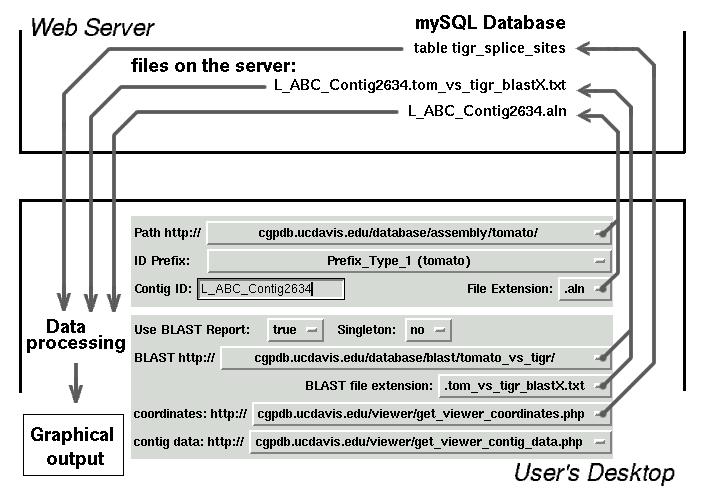
Two PHP scripts
get_viewer_contig_data.php
and
get_viewer_coordinates.php
should be placed into defined web directories.
Those PHP scripts interact with mySQL database and extract information from
two tables. First table contains EST sequences. Second table contains data
for splicing sites of Arabidopsis thaliana. Details about tables set up can
be found at:
CGPDB mySQL.
Information about splicing sites for Arabidopsis genome was derived using Python
GenBank Parser.
Note, that drawing of splicing sites does not take into consideraration
gaps on BLAST alignments. It means, if gaps are large, position(s) of introns
may be slightly shifted to their actual position(s).
BLAST reports for every sequence or contig also should be placed into pre-defined web
directories. Path to those directories is embedded into the source code of Contig Viewer.
Proper set up of scripts and database requires moderate level of experince in mySQL
and PHP. Questions and comments are very welcome: akozik@atgc.org, Alexander Kozik.
