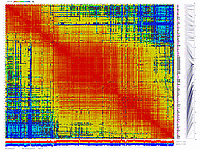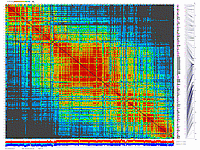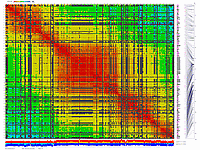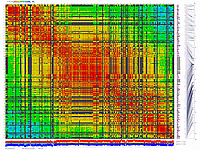

Python_MadMapper_V248_RECBIT_012.py checks all possible
combinations of "triplets" of markers for double crossovers and generates output with "best marker pairs/triplets".
It works in cubic time, NxNxN, where N is a number of markers. Script generates a list with possibly noisy (bad) markers.
Read more about this 248 version README_MADMAPPER_248
Example test locus file: madmapper_scores.loc
Python_MadCluster_V224_GENERAL.py is designed to work
with "general" binary data to cluster nodes. It uses "00", "01", "10" and "11" values to create distance matrix
and following clustering.
Python_MadMapper_V112_RECBIT.py was written for CGPDB project
to assist in construction, validation and visualization of
lettuce genetic map.
Arabidopsis data were used to check program functionality and compare results with lettuce recombination data.
email to: akozik@atgc.org Alexander Kozik
last modified August 29 2005