UC Davis
Rationale
ESTs, NCBI definition:
Expressed Sequence Tags (ESTs) are short (usually about 300-500 bp),
single-pass sequence reads from mRNA (cDNA). Typically they are produced in large batches.
They represent a snapshot of genes expressed in a given tissue and/or at a given developmental stage.
They are tags (some coding, others not) of expression for a given cDNA library.
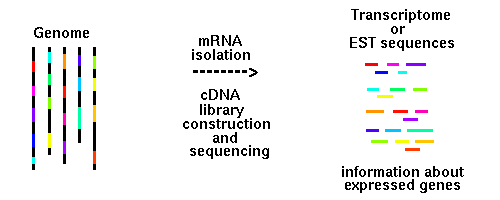
Why we do cDNA library construction and ESTs sequencing:
Plant genomes are large. Whole genome sequencing is expensive. Genome assembly and gene prediction
programs are far away from perfection.
Alternative way to study plant genome content is a sequencing of cDNA clones. This approach can
reveal a substantial portion of the expressed genes of a genome at a fraction of the cost of genomic sequencing.
Largest plant ESTs sequence collections are available at
TIGR Gene Indices database.
Wide variety of species are represented at TIGR database such as maize, rice, wheat and others. Our contribution to
this collection was generation of more than 120,000 lettuce and sunflower ESTs.