UC Davis
Clustering analysis by Graph9 program.
BLAST EST assembly against itself -->
--> Generation of "Matrix" file using tcl_blast_parser.tcl program -->
--> Clustering and bridges search by Graph9 program.
Graph9 output with bridges info, see table lettuce_clustering at CGPDB:
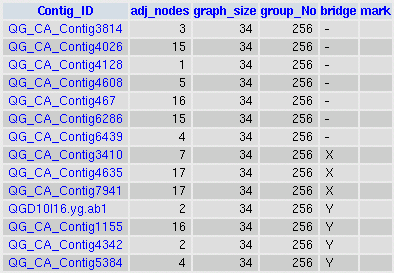
Clustering parameters were: Expect cutoff 1e-20, Identity cutoff 40% and Overlap cutoff 100 amino acids.