UC Davis
Step 9: Processing of the CAP3 output
Obtaining of the "Contig_Info" file
At this step we have used Python_CAP3_ContigExtractor_Feb_27_2004.py
script with option "1" to process CAP3 output and generate so called "Contig Info"
file. Below is the example of Python_CAP3_ContigExtractor_Feb_27_2004.py dialog:
$ python Python_CAP3_ContigExtractor_Feb_27_2004.py
What type of output do you want? (1/2): 1
Enter the SOURCE file name: tomato_ABC.cap3.out
Enter the DESTINATION file name: tomato_ABC.cap3.out.Info
tomato_ABC.cap3.out.Info file has been generated.
Below, is an example of five lines taken from
tomato_ABC.cap3.out.Info file:
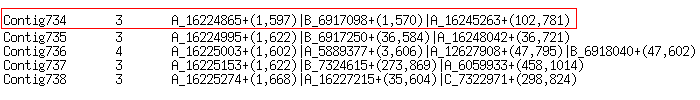
Selected Contig734 contains three ESTs in the assmbly and has length of 781 nucleotides.
Regions of spanning for every EST in the contig are displayed in brackets.