UC Davis
Step 11: Processing of the "Alignment Files" and
finding all mismatches in ESTs versus consensus sequences
At this step we have used Python_CAP3_MM_Finder_Feb_27_2004.py script to
find all mismatches in ESTs versus consensus sequences.
We need to change working directory to "tomato_alignments" and combine all "Alignment Files" into
one file "L_ABC_Contigs_All.aln"
using UNIX cat.
$ cd tomato_alignments
$ cat L_ABC_Contig* > L_ABC_Contigs_All.aln
Execute Python_CAP3_MM_Finder_Feb_27_2004.py script in that directory.
Example dialog is shown below:
$ python Python_CAP3_MM_Finder_Feb_27_2004.py
Enter the SOURCE file name: L_ABC_Contigs_All.aln
Enter the DESTINATION file name: L_ABC_Contigs_All.aln.MM
L_ABC_Contigs_All.aln.MM
file will be generated containing
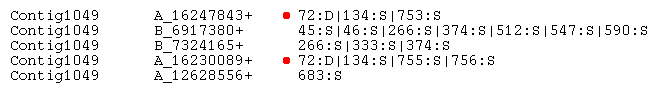
information about all polymorphic sites. Example lines for the Contig1049 are shown below.
"D" is deletion, "I" - insertion, "S" - substitution. Compare this information for
position "72" [red dot] to the alignment in the previous step.