UC Davis
Step 18: Oligo design
Final steps of our SNP/INDEL pipeline are oligo (primer) design for PCR based
molecular markers.
We are using Primer3 program and
contig_target_May_01_2003.pl Perl script
to generate a set of oligos (primers) for PCR.
contig_target_May_01_2003.pl script takes as an input
two files:
1. Table generated by Py_ContigViewer with info about region of interest
2. Tab-delimited file with consensus sequences
This web page describes briefly how to generate a table with "target" info.
Next web page - how to obtain tab-delimited file with consensus sequences
and "step 20" - how to run contig_target_May_01_2003.pl
script and description of its output.
Detailed description of Py_ContigViewer is available
here. Below,
just example screenshots for three selected contigs and table with "target" info
generated by the viewer:
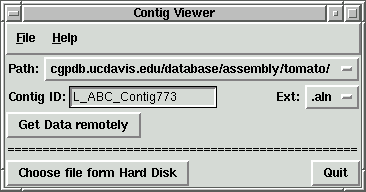
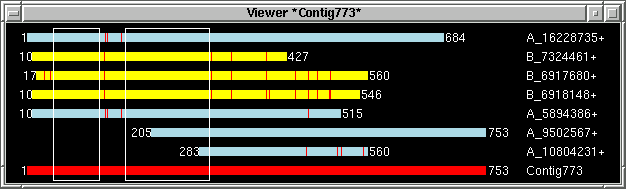
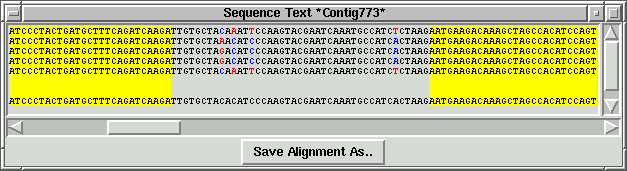
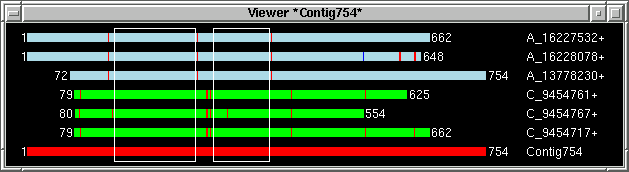
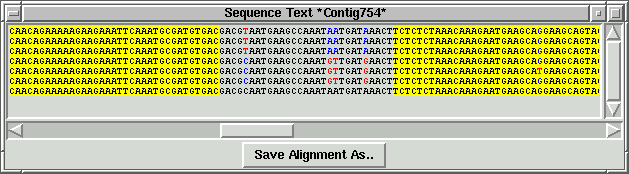
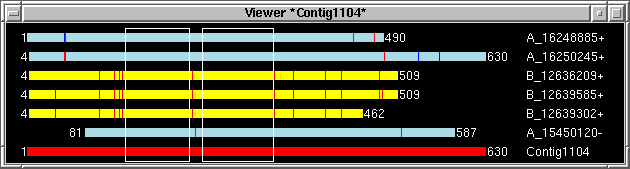
By using "Add to list" function "primer_picker.in"
table has been generated with "target" info for three selected contigs.